Recent research reveals a relationship between sleep disturbances, the orexinergic system, and cognitive decline in Alzheimer’s Disease (AD), highlighting the potential of orexin receptors 1 and 2 (OX1R and OX2R) as promising new targets for AD therapeutic intervention.
In this project, we have used virtual screening and structure-based drug design to identify potential molecular photoswitches for OX1R and OX2R. Photoswitches undergo conformational changes upon light exposure, potentially altering their pharmacological effects. This offers a way to regulate orexin receptor activity in a specific and reversible manner, providing a powerful research tool for studying the orexin receptor's role in AD and other neurodegenerative disorders.
In the initial phase of our study, we focused on curating and refining datasets of azobenzene-containing molecules, a widely used photoswitchable group. Simultaneously, we investigated the structural features of orexin receptors using InfiniSee’s Protein and Binding Site Modes.
In the latest phase of our study, we focused on docking the azobenzene-containing molecules in both their cis and trans isomeric forms to predict their binding affinities and guide the selection of promising candidates for further experimental validation and optimization. Moreover, while these molecules have not yet been evaluated in biological assays, we have explored the potential of structure-based optimization. This entailed validating a protocol for fragment extension and scaffold hopping of known orexin receptor agonists and antagonist scaffolds.
After 1 year, Diana Vicente has achieved the following goals:
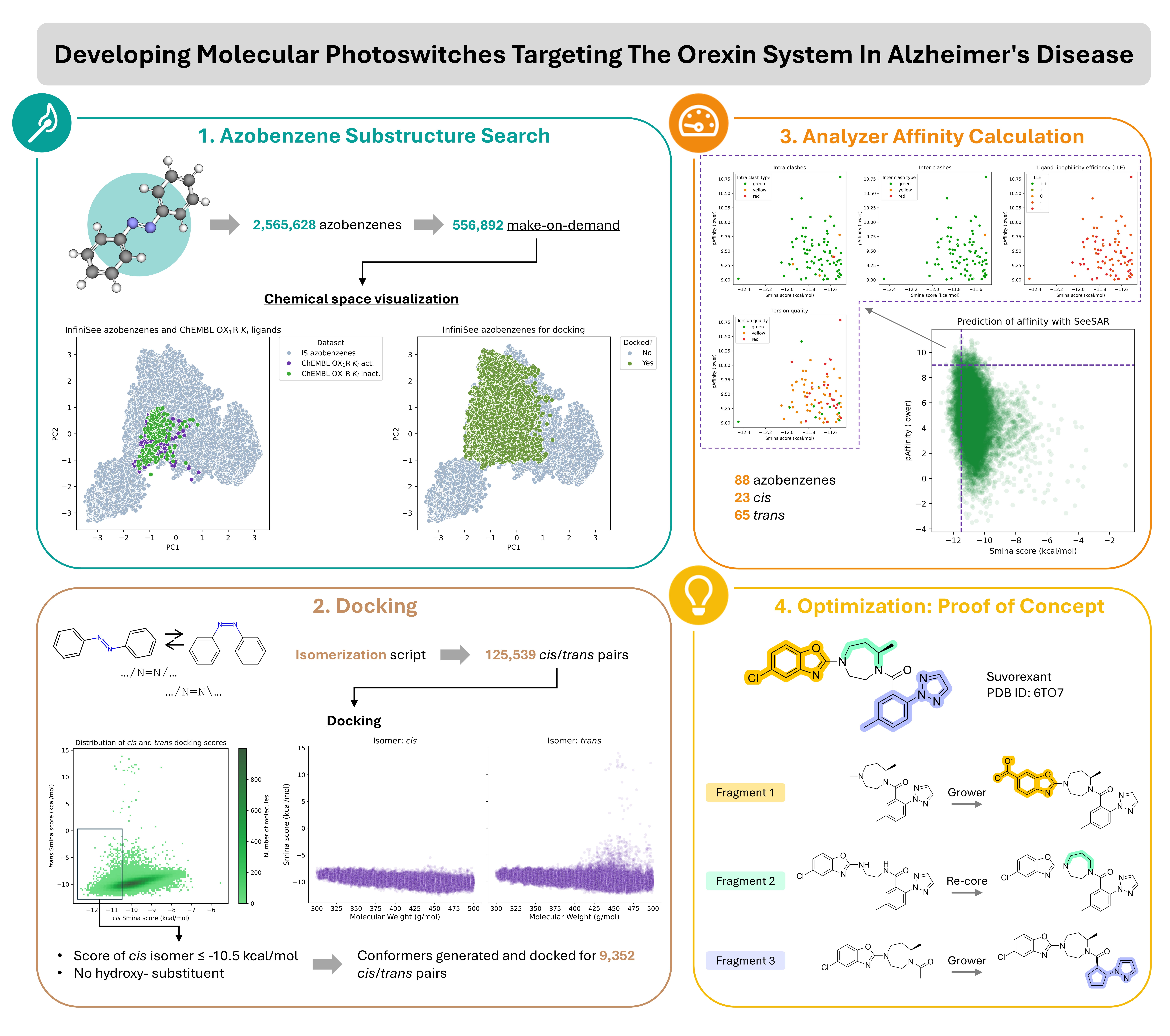
- Using InfiniSee, we performed a substructure search for azobenzenes across all chemical spaces, resulting in 2,565,628 “make-on-demand”, “possible make-on-demand”, and “do-it-yourself” compounds. We focused on the make-on-demand set, narrowing the list to 556,892 azobenzenes. These were compared with known OX1R ligands with Ki data from ChEMBL, using t-SNE dimensionality reduction calculated on the Morgan fingerprints. To refine our selection, we filtered out molecules that RDKit could not process, did not adhere to Lipinski's Rule of 5, or contained PAINS or BRENK substructures (other than the azo group). Finally, we reassessed the overlap with OX1R ligands to ensure relevance for docking studies.
- We generated cis and trans isomers for the azobenzene ligands and prepared them for docking. Hydroxy-substituted azobenzenes were excluded due to their known poor photophysical properties. Using an in-house protocol, we docked a single conformer of each ligand into OX1R. We then analyzed the correlation between the docking scores and molecular weight, as well as the correlation between the docking scores of the cis and trans isomers. For those ligands with a cis isomer docking score below -10.5 kcal/mol, we generated 30 conformers and docked them again. The final poses were uploaded to SeeSAR, where we used the Analyzer Mode to estimate the binding affinity, the torsional quality, intra- and inter-molecular clashes, and pharmacokinetic properties.
- We conducted a preliminary study where we generated fragments of the well-known dual orexin receptor inhibitor suvorexant and used the Inspirator Mode to attempt to reconstruct the original molecule within the binding site of the receptor (PDB ID: 6TO7). This was done to validate the protocol for future optimization of our photoswitches. We evaluated the results by identifying the Inspirator-derived molecule with the largest maximum common substructure to the starting molecule. The results were successful in recovering molecules similar to suvorexant, demonstrating the applicability of this approach for future optimization efforts.