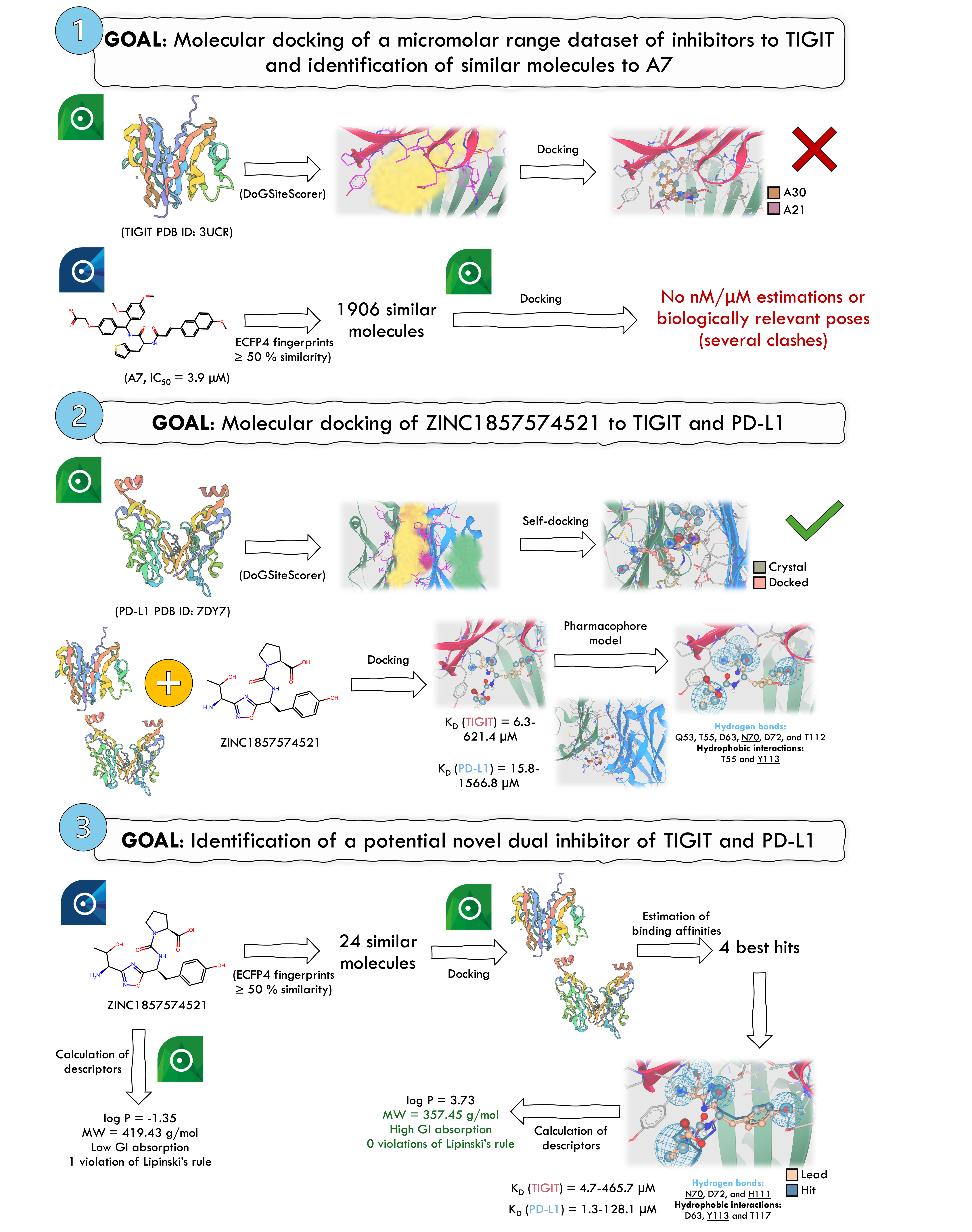
TIGIT is an immune checkpoint protein that when overexpressed in NK and T cells is associated with poor cancer prognosis. Currently, there are no small inhibitors targeting TIGIT. In this study, we aimed to identify potential inhibitors of TIGIT. We used SeeSAR to select a TIGIT structure(3UCR) and identified its binding pocket via DoGSiteScorer. Initially, we evaluated a dataset from Xiong et al., comprising 14 active and 21 inactive compounds, through docking that involved generating 500 poses with medium clash tolerance. However, no poses were generated for 25 compounds, including the most potent one, A7(IC50=3.9 µM). Hence, we explored compounds similar to A7, employing a 50% similarity threshold via infiniSee. However, the generated poses presented numerous clashes. Therefore, we adopted an alternative strategy, selecting ZINC185757452 from the WO2019175799A2 patent, in which 1,2,4-oxadiazole compounds derivatives are described as TIGIT/PD-L1 inhibitors. We uploaded the TIGIT and PD-L1(7DY7) structures and identified their binding pockets. Self-docking of the PD-L1 ligand confirmed SeeSAR’s accuracy (KD=25 nM, estimated range 0.3-32.7 nM), with an RMSD of 3.52Å. This discrepancy was attributed to the solvent-exposed region, although the non-exposed areas aligned well visually. Docking studies with ZINC1857574521 revealed estimating KD values of 6.3-621.4µM(TIGIT) and 15.8-1566.8µM(PD-L1). A visual inspection identified key interactions, but confirmation of hydrophobic contacts required further analysis using additional softwares. Key interactions involved H bonds with residues Q53, T55, D63, N70, D72, and T112 of TIGIT and hydrophobic (Hyd) interactions with T55 and Y113. N70 and Y113 were previously reported as crucial. To refine our search, we generated a pharmacophore featuring 1 Hyd site, 1 acceptor(Acc)/donor(Don), 1 Acc, and 2 Don. Using infiniSee, we identified 24 compounds with ≥50% similarity to ZINC1857574521. Docking results revealed 4 compounds with greater KD than ZINC1857574521. The most promising candidate from REALSpace database exhibited a KD=4.7-465.7µM and 50.7% similarity to ZINC1857574521. It demonstrated improved ligand efficiency, fewer intramolecular clashes, and fulfilled 3 pharmacophoric features, forming key interactions with TIGIT (N70, H111 – H bonds; Y113 – Hyd) alongside D63, T117, and D72. Additionally, docking to PD-L1 gave an estimated KD of 1.3-128.1µM, surpassing the initial compound. Chemical descriptors were calculated using SeeSAR, and other software predictions deemed the hit as "drug-like" and soluble. In conclusion, we identified a promising TIGIT/PD-L1 inhibitor, which can revolutionize cancer treatment as co-inhibition strategies are prone to elicit more significant anticancer responses.
After 1 year, Rodrigo has achieved the following goals:
- Molecular docking of a µM-range dataset of inhibitors to TIGIT and identification of similar molecules to A7 In SeeSAR’s software, we uploaded 1 TIGIT structure (PDB ID:3UCR), and then we used DoGSiteScorer to choose the binding pocket region (score=0.48). After this, crucial amino acid residues were de/selected according to the literature. A small dataset ofµM range inhibitors from Xiong et al. with 14 active and 21 inactive compounds was first used. A maximum of 500 poses and a medium clash tolerance were selected, but no poses could be generated for 25 molecules, including active ones such as the most active(A7,IC50=3.9 µM). Plus, compounds predicted to bind with the higher affinity were inactive. This approach failed to generate biologically relevant poses, so we looked for similar compounds to A7 instead. A similarity threshold of 50% was used, and then hits were docked to TIGIT. Then, affinities were estimated, but none was within the nM/µM range, and poses had several clashes.
- Molecular docking of ZINC1857574521 to TIGIT and PD-L1 As a backup plan, we used the exemplifying compound from the WO2019175799A2 patent (ZINC1857574521). Then, TIGIT (PDB ID: 3UCR) and PD-L1 (PDB ID: 7DY7) were uploaded to SeeSAR’s software, and respective binding pockets were selected using DoGSiteScorer with scores of 0.48 and 0.46, respectively. Validation of the PD-L1 structure was made by performing self-docking. Molecular docking to PD-L1 and TIGIT was performed, binding affinities were estimated, and poses were visually inspected. For TIGIT, ZINC1857574521’s KD estimation was 6.3-621.4 µM and, for PD-L1, was 15.8-1566.8 µM. Then, from a visual inspection, the main interactions involved with TIGIT were identified, but hydrophobic ones had to be confirmed by an external software. Based on the docked complex, a pharmacophore model was generated. It included 5 features: 1 hydrophobic (with a 2.0 Å radius), 1 acceptor/donor, 1 acceptor, and 2 donors (default radii).
- Identification of a potential novel dual inhibitor of TIGIT and PD-L1 We searched for molecules with at least 50% similarity to ZINC1857574521 using ECFP4 fingerprints with infiniSee. 24 compounds were identified from AMBrosia, eXplore, and REALSpace, and then docked to TIGIT. 4 were predicted to have better affinity. The compound predicted with the best binding affinity to TIGIT (4.7-465.7 µM) from ENAMINE (50.7 % similarity) showed a better ligand efficiency and could also meet 3 features of the generated pharmacophore. Plus, it could establish interactions with TIGIT that had been seen as key. Moreover, this compound was also docked to PD-L1, and the predicted KD was 1.3-128.1µM. Finally, we calculated some descriptors using SeeSAR and saw that this compound was drug-like and soluble using an external software. In conclusion, we were able to identify a hit that may be of great interest for dual inhibition of TIGIT and PD-L1 and can pave the way in cancer treatment.