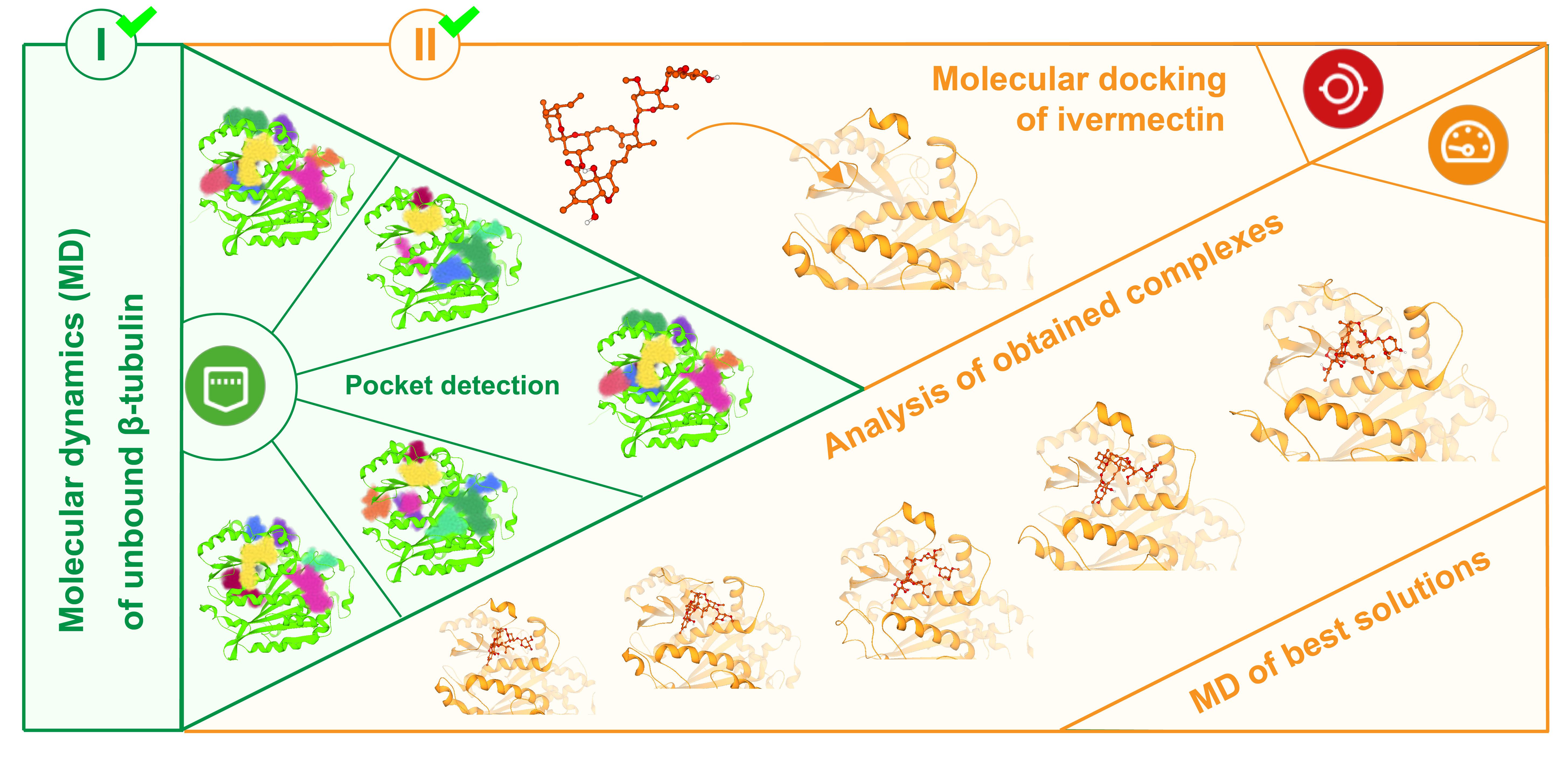
During the first three months of the project two of the three milestones were reached. Particularly, the conformations of unbound β-tubulin of M. enterolobii, M. graminicola, M. hapla, M. incognita, and C. elegans were collected using molecular dynamics and the obtained trajectories containing the conformations were analyzed. The major conformational states of each studied structure were determined and extracted from the trajectories. Using the Binding Site mode of SeeSAR pockets on the surfaces of extracted confromations were detected, analyzed, and filtered based on the volume and shape. Further, the molecular docking of ivermectin to each selected conformation was performed and thus the complexes of ivermectin and tubulin were obtained and analyzed in SeeSAR. Finally, the molecular dynamics of best complexes were performed. The further analysis with SeeSAR and PoseView is necessary to reach the Milestone 3.
After 3 months, Yevhen has achieved the following milestones:
- Classical molecular dynamics simulations of unbound β-tubulin of M. enterolobii, M. graminicola, M. hapla, M. incognita, and C. elegans were performed using GROMACS. The cluster analysis of collected conformations of each target was done and the middle conformations of determined clusters that occur with high frequency were identified and extracted from the trajectories. Binding pockets were further detected and analyzed in the SeeSAR’s Binding Site mode. Conformations of the predicted binding site with optimal shape and volume were selected for the molecular docking.
- Molecular docking of ivermectin to the predicted binding site was performed in the Docking mode of SeeSAR for selected conformations of each studied structure. The obtained complexes of ivermectin and β-tubulin were analyzed in the Analyzer mode of SeeSAR. Consequently, from each structure’s pool of complexes the best was selected for simulations based on affinity and geometrical properties. Finally, the molecular dynamics simulations of selected complexes were performed.
- The analysis of molecular dynamics trajectories containing conformations of complexes has been done. However, the further analysis of collected conformations with SeeSAR and PoseView is required to determine SAR of IVM-tubulin complexes and to highlight perspectives of further research.