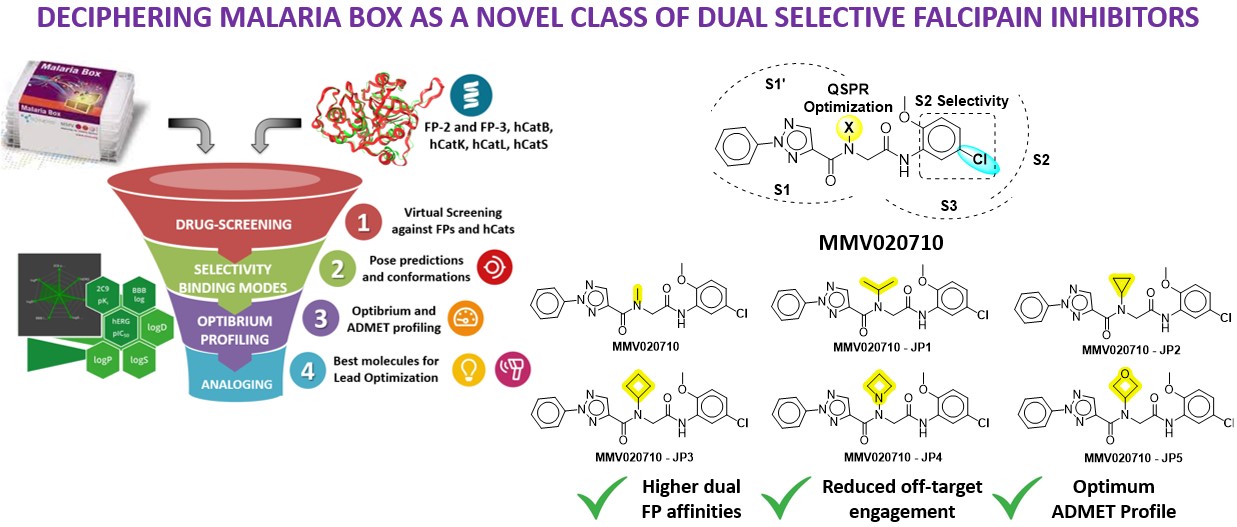
To begin with, I’d like to express my gratitude to BioSolveIT software in view of the remarkable speed, quality, and elegance that made the year productive. My proposal was related to my PhD theme of exploring a new class of antimalarial regimens that could selectively inhibit Falcipain. However, I was unable to land on an appropriate platform to validate my hypothesis. Soon, the Fall 2023 Scientific Challenge came on my way to give life to my project. To combat drug resistance, the identification and validation of new bioactive molecules are crucial for accelerating parasite reduction. The research builds on the Malaria Box, a collection of potent antimalarial candidates by the Medicines for Malaria Venture, Switzerland. These compounds are effective across various stages of the parasite life cycle. Here, drug repositioning and optimization against dual falcipain (FP2/3), aim to reduce off-target engagement with highly homologous human cathepsin isoform. Using the BLOSUM62 matrix, we retrieved key PDB ID 3BPF (FP-2), 3BWK (FP-3), and 4DMY (hCatK). Further, the MMV library against all proteins, identified 150 hits with picomolar to nanomolar binding affinities. These candidates were further analyzed against hCatK, where the top 30 hits exhibited micromolar affinities. The S2 subsite, characterized by hydrophobic residues and smaller groove sizes in FP proteins, emerged as a critical determinant for selectivity. MMV020710 stood out for its potent activity against FPs and minimal off-target interaction, attributed to the presence of a -Cl at the meta position, enhancing hydrophobic contacts with the S2 subsite of FP proteins while avoiding hCatK. Despite its promising profile, MMV020710 required improvements in solubility and permeability, which were addressed through structural analoging. In the final milestone, we performed structural analoging on MMV020710, focusing on the methyl substituent, which is solvent-exposed. By designing analogs with randomization at this site, we aimed to optimize quantitative structure-property relationships (QSPR). Modifications at other sites were avoided to maintain selectivity against hCat isoforms. Toxicophoric fragments and those under PAINS and REOS categories were excluded. Incorporating solubility-enhancing groups such as gem-dimethyl and monocyclic rings led to significant improvements in solubility and permeability. The resulting MMV020710 analogs exhibited 1/800 times inhibition for FPs over hCatK. Further, frontier molecular orbital was done to gain deeper insights into the reactivity and selectivity. The newly designed molecules underwent alchemical-steered simulation to confirm their inhibition. Synthesis and biological evaluation of the best hits can result in successful outcomes in the future.
After 1 year, Jeevan has achieved the following goals:
- To overcome drug resistance, identifying and validating new bioactive molecules can expedite the parasite reduction in the intra-erythrocyte stage. Previously, the Malaria Box was designed by the Medicines for Malaria Venture (MMV), Switzerland having highly potent antimalarial clinical candidates. These candidates are proven to act at various stages of the parasite life cycle. In this study, drug repositioning against FP was studied which also could possess lesser off-target engagement against highly homologous hCat isoforms. From the BLOSUM62 matrix using falcipain sequence, we retrieved PDB 3BPF (FP-2), 3BWK (FP-3), and 4DMY (hCatK) from the PDB bank and geometrically optimized them.
- The MMV library was screened against dual FP-2 and FP-3 proteins. Among them, 150 molecules exhibited a nanomolar binding affinity range against dual FPs. These were further analyzed with hCat isoforms in which the top 30 hits were shown at the micromolar range. The S2 subsite of all proteins is the key site for achieving selectivity. It possesses hydrophobic residues and smaller groove sizes in FP proteins. Based on the S2 residual contacts, affinities, and optibrium properties MMV020710 has no off-target engagement with hCat and is highly potent towards FPs. This is likely due to the presence of -Cl (EWG) at the meta position has higher hydrophobic contacts projecting towards S2 while lacking hCatK protein. However, there is still a need for solubility and permeability enhancement of MMV020710 which were achieved through structural analoging.
- The structural analog fragments using SpaceLight, SpaceMACS, and CoLibri were performed on the identified MMV020710 at methyl substituent. Because the methyl group is projecting towards the solvent exposure area. Designing analogs upon randomization on methyl group could be a niche for QSPR study. Analogues at other sites are much likely to interact with hCat residues which may fail to achieve selectivity. Fragments that are toxicophores and under PAINS (Pan Assay Interference), and REOS (Rapid Elimination of Swill), categories are eliminated from the analogue series. Substituting solubility enhancers such as gem-dimethyl, and monocyclic rings such as cyclopropane, cyclobutane, azetidines, and oxetanes gave promising solubility and permeability improvements. The MMV020710 analogs have two-fold higher affinity with dual FPs over hCat isoforms. These newly designed molecules were further validated using conventional simulations and alchemical free-energies methods.