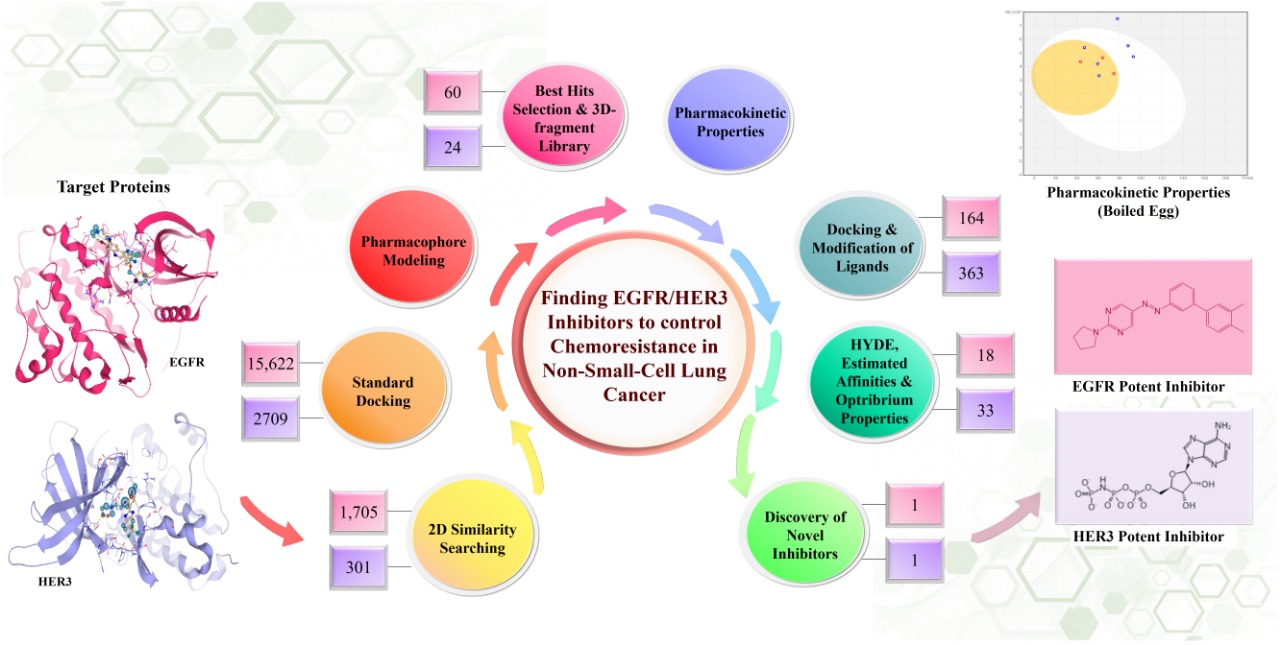
Lung cancer is the most intermittently diagnosed malignancy with a high fatality rate across the globe. Among lung cancer, approximately 85% of the patients suffer from non-small-cell lung cancer (NSCLC), a heterogeneous disease characterized by numerous molecular aberrations. Despite innumerable scientific research and therapeutic strategies, the probability of survival in advanced stage NSCLC is still obscure. The foremost obstacle is the mutations in molecular targets that renders the therapy ineffective and develops chemoresistance. Therefore, the project was designed to construct inhibitors for targeting proteins to resist chemoresistance. HER3, also known as ERBB3, was preferably selected because of its specificity to bind with a single ligand and role in therapy failure and enhanced cell survival. Along with HER3, epidermal growth factor receptor (EGFR) was also targeted. To achieve the synthesis of potent inhibitor, BioSolveIT tools and softwares were used. The targeted proteins and their active pockets, similar compounds for inhibitors, molecular docking, pharmacophore modeling, hits generation and screening, molecular editing, pharmacokinetics, and optribrium properties were all analyzed using these softwares. During the project, the ReCore in inspirator mode and molecular editor mode in SeeSAR, being user friendly, have exceptionally assisted in designing a potent inhibitor.
After 1 year, Imtiaz has achieved the following goals:
- The project was initiated by finding reported inhibitors from literature for the targeted proteins. These inhibitors were screened on the basis of binding affinities with the targeted proteins subsequent to standard docking via SeeSAR. Afterwards, the screened reported inhibitors were used to search similar compounds from build in chemical space libraries in InfiniSee using FTrees. These compounds were exported and saved to form specific compound libraries for each of the proteins.
- The binding site of the proteins were expanded by adding more residues at the active pocket in binding site mode in SeeSAR to facilitate the docking analysis. Subsequently, all the similar compounds for each of the target proteins were docked using FlexX (SeeSAR) and were analyzed on the basis of their optribrium properties for virtual screening. The screened molecules were employed for developing pharmacophore, and then exported for improvement via inspirator mode. In the respective mode, the estimated affinities, torsions, clashes and LLE were ameliorated with the help of fragment growing and core-replacement properties.
- The screened molecules were rendered as hits and were screened for pharmacokinetic and druggable properties using SeeSAR and SwissADME. Some molecules gave remarkable results and showed all the druggable properties. On the other hand, a few molecules were also quite good but needed some corrections; therefore, they were amended and refined by molecular editor mode. This mode is enthralling for a chemist and incredibly enhance the features of a molecule to be a potent inhibitor. The improved molecules were further analyzed for optribrium and pharmacokinetic properties. Ultimately, the potent inhibitors for both the target proteins were synthesized for in vitro analysis.