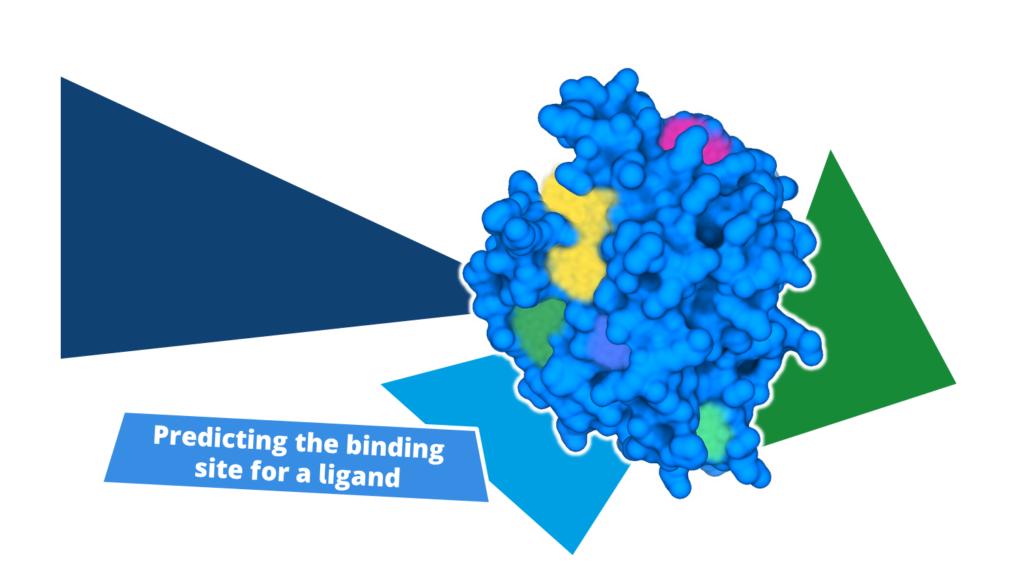
To validate your prediction, it’s essential to combine information on ligand activity with insights into its interactions with the target structure.
Dock your known actives into the predicted binding site and assess the predicted binding modes to rationalize structure-activity relationships (SARs). Having a series of compounds to work with significantly simplifies the process.
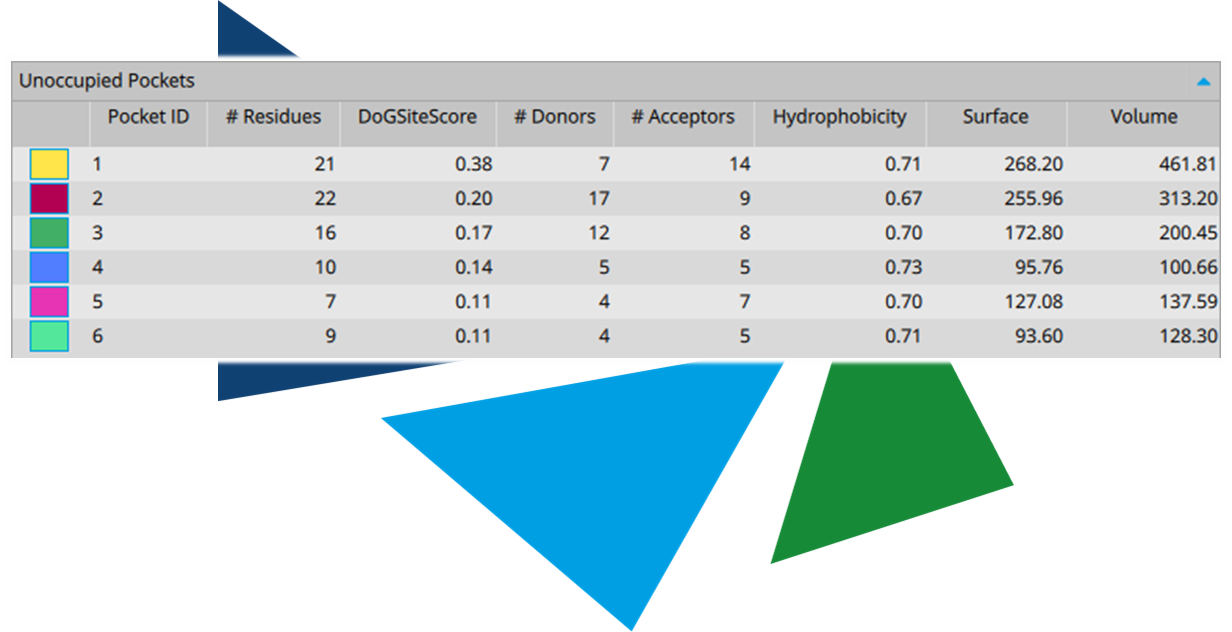
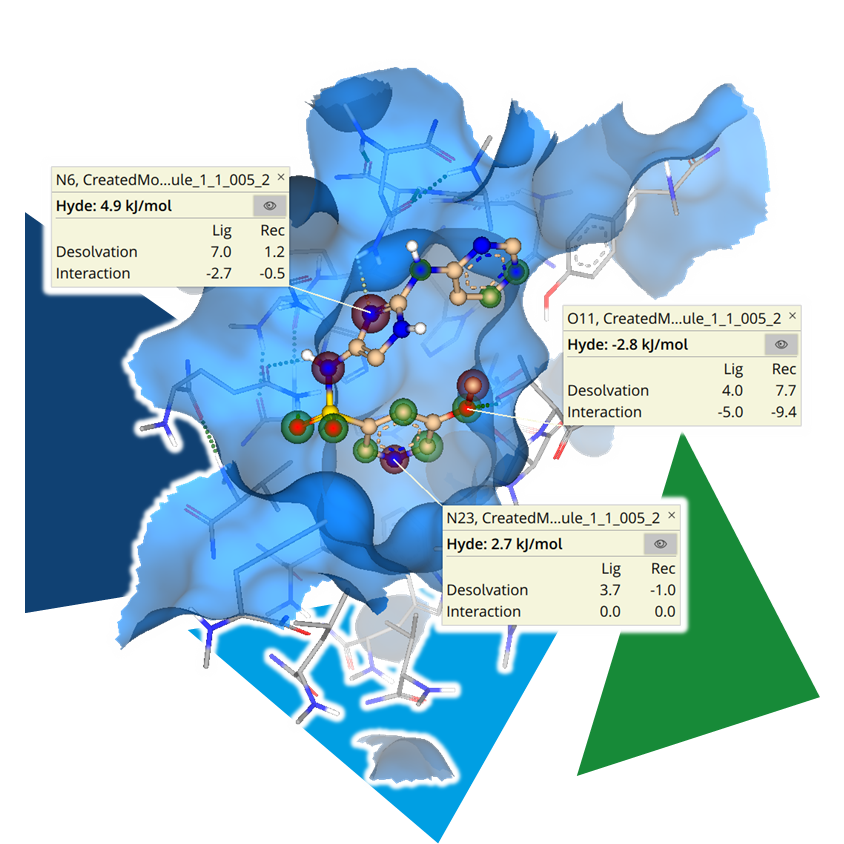
Important functionalities of the compounds should bind in areas where they are tolerated or, ideally, form favorable interactions. Our scoring algorithm
HYDE helps you spot the individual contributions of each heavy atom, as well as the quality of the formed H-bonds. Build up on gained insights to iterate your initial hypothesis and adjust accordingly.