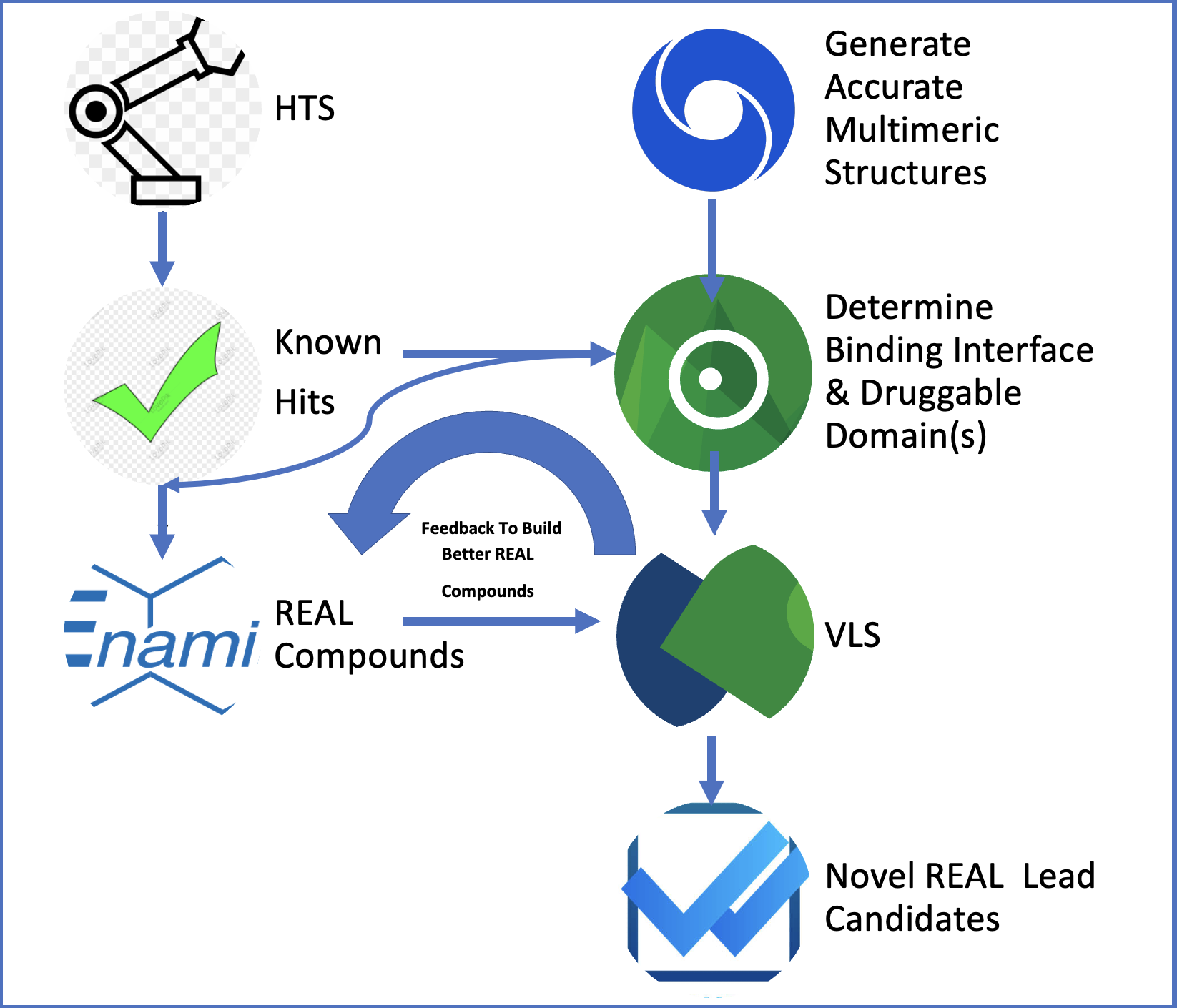
Over the past 12 months, our project 'REAL VLS to Find Activators of BMP Signaling using Neural Network Crystal Structures' has made significant progress toward identifying novel BMP7 mimetics that can bind to the ALK3 receptor, potentially stimulating the regeneration of beta cells from progenitor cells. This journey has been greatly facilitated by the comprehensive tools and suites provided by BioSolveIT, particularly SeeSAR and infiniSee.
infiniSee has been instrumental in enabling us to explore a vast chemical space efficiently. By leveraging its advanced algorithms, we were able to search through millions of potential ligands that could act as BMP7 mimetics. This large-scale virtual screening was crucial in narrowing down the candidate compounds that exhibit the desired binding properties with the ALK3 receptor. The software’s ability to handle massive datasets and provide quick, reliable results was essential in accelerating our initial discovery phase.
Using the powerful docking and scoring functionalities of SeeSAR, we conducted extensive in silico assays to predict the binding affinities and modes of interaction between the potential ligands and the ALK3 receptor. SeeSAR's intuitive interface and robust features, such as the HYDE scoring system and ADME property assessments, allowed us to make informed decisions quickly. We identified several promising hits through these assays, which were then prioritized for further in vitro testing.
The hits identified from our in silico assays were subjected to rigorous in vitro testing. We were able to validate a few potential hits that demonstrated significant activity in stimulating BMP signaling. These compounds are currently being scrutinized and developed further through additional biochemical and cellular assays to confirm their efficacy and safety.
The automatic binding site detection feature was particularly helpful in identifying the key interaction sites on the ALK3 receptor. We expanded the binding site using additional residues to find potential empty pockets for ligand binding.
Molecule Editor and Analyzer Modes:
The integration of BioSolveIT's infiniSee and SeeSAR tools into our workflow has significantly streamlined our research process. The ability to efficiently screen vast chemical libraries, predict interactions, and modify compounds dynamically has been pivotal in our progress. We are now poised to further develop our identified hits, thanks to the robust and versatile capabilities provided by BioSolveIT’s software. We believe that these tools will continue to be invaluable as we advance towards our goal of discovering effective activators of BMP signaling for beta-cell regeneration.
After 1 year, J. Michael has achieved the following goals:
- Goal: We successfully identified the binding sites on the ALK3 receptor for potential BMP7 mimetic ligands without requiring a crystal structure. Explanation: Using BioSolveIT's advanced tools, particularly SeeSAR, we leveraged the protein mode to load and prepare the ALK3 receptor for analysis. The software's binding site mode automatically detected potential binding pockets, which we then expanded and refined. By utilizing the molecule editor and docking modes, we screened numerous ligands and assessed their interactions with the identified binding sites. This allowed us to predict binding affinities and modes of interaction effectively, even in the absence of a crystal structure. The HYDE scoring system and ADME property assessments further aided in filtering and prioritizing the most promising candidates. This achievement underscores the powerful capabilities of SeeSAR in drug discovery, facilitating accurate predictions and efficient identification of key binding interactions.
- Goal: We saved substantial time and resources by using virtual screening methods, eliminating the need to purchase and test as many compounds as required for a traditional high-throughput screening (HTS) assay. Explanation: By utilizing BioSolveIT’s infiniSee and SeeSAR software, we were able to conduct comprehensive virtual screenings of millions of potential ligands. This approach allowed us to efficiently filter and identify the most promising candidates without the need for extensive physical compound libraries. The in silico assays performed using SeeSAR provided detailed insights into binding affinities and ADME properties, significantly reducing the number of compounds that needed to be synthesized and tested in vitro. This virtual screening process not only accelerated our research timeline but also resulted in considerable cost savings, as fewer reagents and materials were required. Consequently, we were able to allocate our resources more effectively towards the further deve
- Goal: We successfully identified several ligands that demonstrated promising activity in vitro, which are now being further explored for their potential therapeutic applications. Explanation: Leveraging BioSolveIT's SeeSAR and infiniSee, we conducted thorough in silico screenings and assays to predict potential ligands' binding affinities and modes of interaction with the ALK3 receptor. The hits identified through these virtual methods were synthesized and subjected to rigorous in vitro testing. Several ligands exhibited activity in stimulating BMP signaling pathways, confirming the predictions made during the computational phase. These promising candidates are now undergoing additional biochemical and cellular assays to further evaluate their efficacy and safety profiles.