FastGrow now enables efficient and lightning-fast structure-based peptide design with two new libraries. These are the largest sets to date, containing over 661,000 combinations of proteinogenic amino acids and amide bioisosteres, which can be screened within seconds for the best shape-complementary solutions.
How are the sets designed?
Both sets are based on 20 proteinogenic amino acids linked through amides and their bioisosteres. Each FastGrow extension begins with an amide/bioisostere, followed by an amino acid, another amide/bioisostere, and finally a second amino acid. The C– and N-termini are translated into an uncharged form (-CONHCH3 and -NHCOCH3, respectively), enabling further extensions and to avoid scoring bias due the charges.
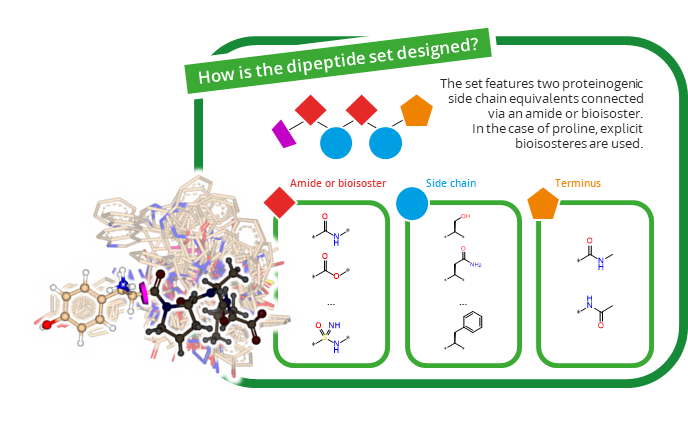
In total, 41 amide bioisosters, together with the amide group, were used in the generation of the library (totaling in 42 functionalities to connect the proteinogenic amino acids). To take into account that not all amide bioisosters can be applied on the proline amino acid, 36 reported proline bioisosters together with the proteinogenic version were used for the functional outliner (totaling in 37 proline-related structures).
Two sets (C– and N-terminus) were generated to enable extensions in both directions of the peptide. Please select the right set when placing your extension vector on your ligand. Here are some examples:
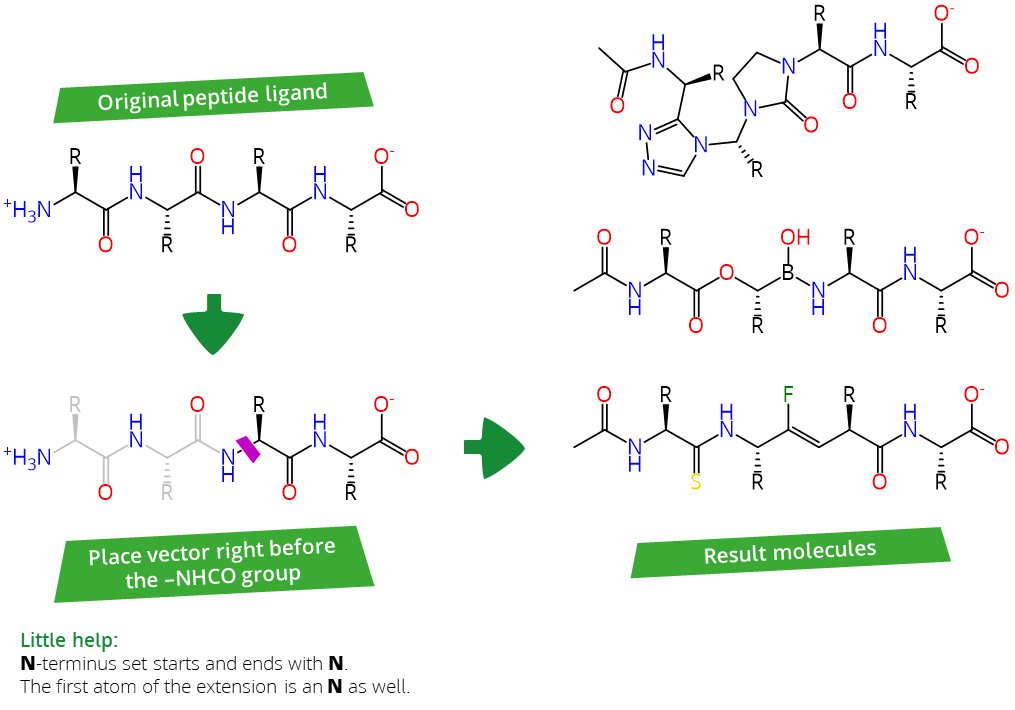
- The N-terminus set: Starts by replacing the Peptide-NHCO-R motif and ends with the uncharged N-terminus.
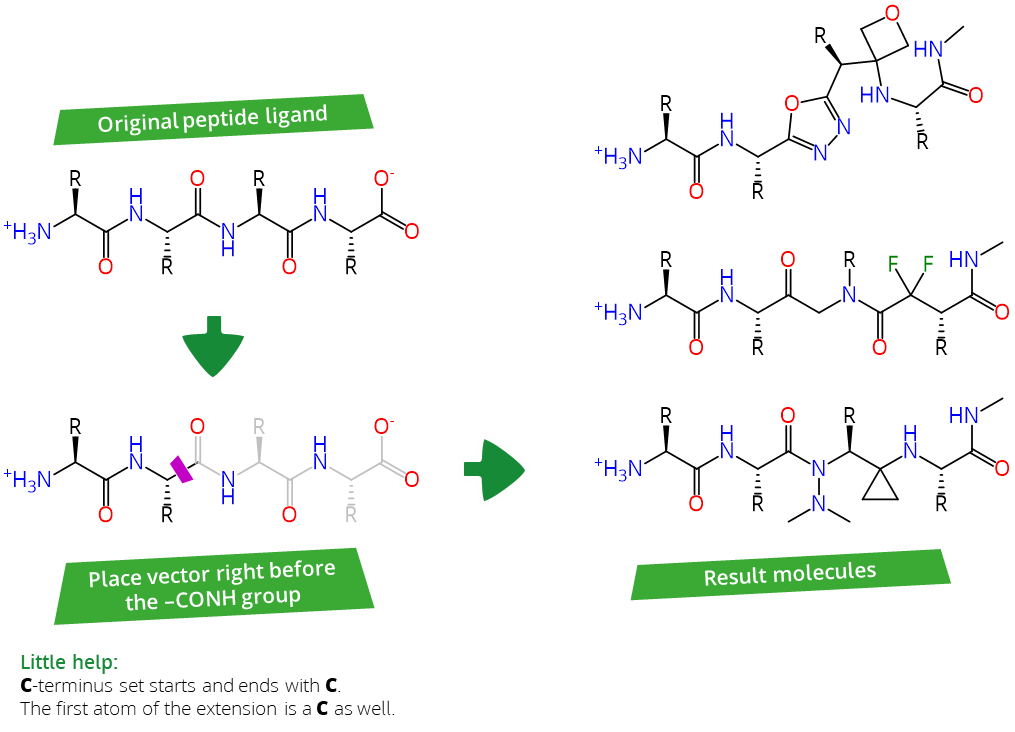
- The C-terminus set: Starts by replacing the Peptide-CONH-R motif and ends with the uncharged C-terminus.
Why Peptide Design and FastGrow are a Perfect Match
Shape and functionality of a peptide are crucial in drug discovery and lead optimization. FastGrow allows the incorporation of pharmacophore constraints to maintain desired functionalities in a ligand, such as preserving specific interaction patterns within a complex. Additionally, our clever programming keeps computational requirements minimized — a critical feature given the high computational demands typically associated with peptides. By factoring in shape complementarity, FastGrow quickly identifies peptide conformations that would clash with the binding pocket, preventing the waste of resources on incompatible configurations — a common issue in docking studies.
Additionally, the use of bioisosteres expands the medicinal chemist’s toolkit. These bioisosteres not only offer metabolically more stable groups compared to amides but also enhance the 3D landscape of the generated fragments. This enables the creation of peptides with improved ADME properties and better filling of the binding pocket, potentially forming additional interactions with residues.
You can download the peptide bioisoster FastGrow libraries following this link.
Some publications on amide bioisosteres:
- Rečnik, L. M.; Kandioller, W.; Mindt, T. L. 1,4-Disubstituted 1,2,3-Triazoles as Amide Bond Surrogates for the Stabilisation of Linear Peptides with Biological Activity. Mol. 2020, Vol. 25, Page 3576 2020, 25 (16), 3576.
- Kumari, S.; Carmona, A. V.; Tiwari, A. K.; Trippier, P. C. Amide Bond Bioisosteres: Strategies, Synthesis, and Successes. J. Med. Chem. 2020, 63 (21), 12290–12358.
- Choudhary, A.; Raines, R. T. An Evaluation of Peptide‐Bond Isosteres. ChemBioChem 2011, 12 (12), 1801–1807.