We are proud to announce not one, but two winners of the Scientific Challenge Winter 2024: Harry Tran and Katharina Buchthal! Due to the exceptional achievements and execution within both projects, it was simply too difficult for us to choose just one outstanding entry. As a result, we decided to honor both with the award.
We wish them both all the best for their careers and look forward to seeing more transformative contributions in the field of drug design!
The project of the first Winner, Harry Tran from the University of Leeds, aims to design novel allosteric inhibitors targeting Mycobacterium tuberculosis DNA gyrase to combat antibiotic resistance. Commonly used antibiotics such as fluoroquinolones are losing efficacy due to resistance mutations. A recently characterized allosteric binding site represent a new mechanism to alter the functionality of the target structure. The binding ligand, the natural product Evybactin, can there be used a new starting point for the development of new inhibitors. His approach combines computational and structure-based workflows, leveraging tools like SeeSAR and FastGrow for de novo drug design.
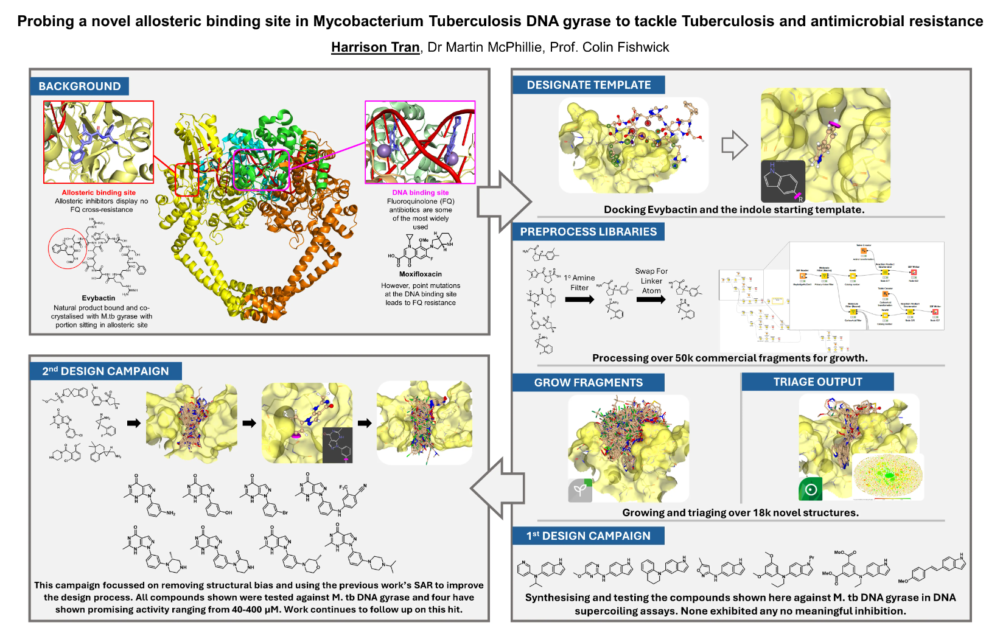
Key Milestones and Achievements:
- De Novo Design:
- Used SeeSAR and FastGrow to create over 18,000 novel molecules across two design campaigns.
- Initial designs were inspired by Evybactin’s indole motif, while the second campaign employed a new template (pyrazolopyrimidinone) to improve potency and synthetic tractability.
- Chemical Synthesis and Evaluation:
- Synthesized 15 novel compounds, with 4 exhibiting promising micromolar activity against M. tb DNA gyrase in supercoiling assays.
- Workflow optimization ensured synthetically feasible outputs, although some scaffolds required extensive chemistry refinements.
- Iterative Design Improvement:
- Biological data from the first campaign informed the second, enabling refinement of structure-activity relationships (SAR).
- Binding affinity predictions improved from micromolar to sub-nanomolar levels, with corresponding increases in biological activity.
Why the project convinced us:
- The project highlights the utility of BioSolveIT’s software suite (SeeSAR and FastGrow) in fragment-based drug discovery and iterative SAR expansion.
- It demonstrated an effective pipeline for exploring allosteric sites, leading to diverse and synthetically accessible inhibitors with significant potential in tuberculosis treatment.
Read more about Harry’s project following this link.
The project of the second Winner, Katharina Buchthal from the Center for Bioinformatics of the Saarland University, focused on developing an automated in silico pipeline to design novel inhibitors targeting PKA kinase, a key player in signal transduction often linked to cancer and other diseases. Using fragment-based drug discovery and KinFragLib, the pipeline generates kinase inhibitors by growing fragments within subpockets of the binding site. Key features include FlexX docking, HYDE scoring, and a cluster-based approach to balance molecular diversity and binding affinity.
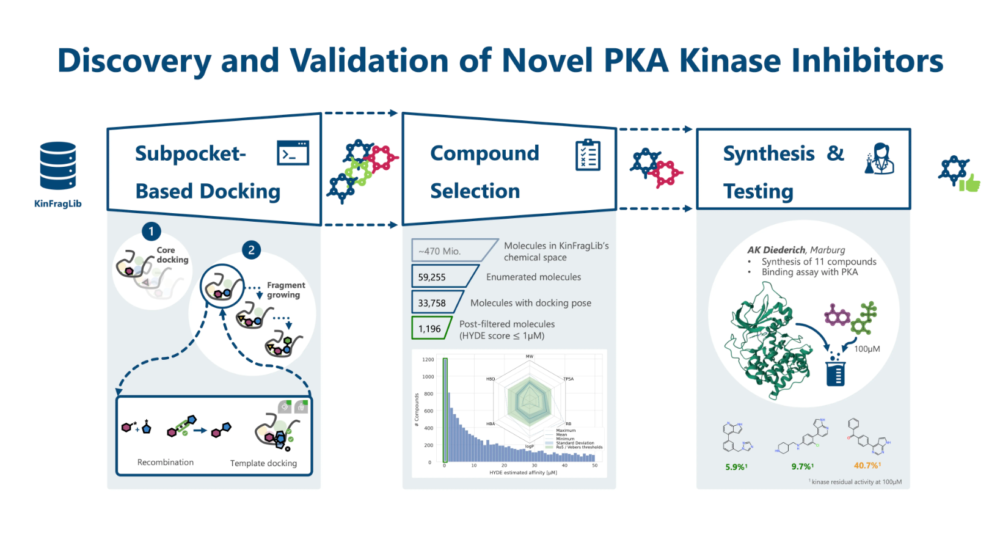
Key Milestones and Achievements:
Pipeline Development and Application:
- Pipeline Development and Application:
- Refined the pipeline for modularity and enhanced molecular diversity, incorporating updated KinFragLib fragments (expanded from 7,486 to 9,505).
- Applied the pipeline to a hamster PKA structure (PDB: 5n1f), generating 33,758 compounds. After filtering, 1,166 candidates with low nanomolar HYDE affinities and drug-like properties were identified.
- Compound Synthesis and Experimental Testing:
- Collaborated with AK Dietrich at Philipps University Marburg for synthesis and validation.
- Of the top 100 prioritized candidates, 16 were selected for synthesis based on diversity and synthetic accessibility.
- 11 compounds were successfully synthesized, with preliminary kinase assays showing:
- 4 compounds reduced kinase activity to <50%
- 2 compounds reduced activity to <10%, all at 100µM concentration.
- Future Refinement and Validation:
- The pipeline is designed to iteratively improve based on experimental feedback. Synthesis outcomes and biological results will guide future designs to enhance potency and selectivity.
Why the project convinced us:
- This work demonstrates the potential of fragment-based pipelines to generate chemically diverse and novel kinase inhibitors. Promising experimental validation highlights its relevance for cancer drug discovery, with the finalized pipeline made available on GitHub for broader application.
Read more about Katharina’s project following this link.





