In the context of our kinase inhibitor webinar, we previously explored how to investigate the chemical space around a compound of interest.
Among those topics, the FastGrow feature of SeeSAR was highlighted as an efficient method to mine for novel hinge binding motifs to complement the orthosteric binding site of the targeted kinase. The underlying algorithm screens a preconstructed library of molecular fragments for the best candidates at ultra-high speed. During the analysis, various conformations of the fragments are considered, and the best solution for the problem is selected.
Now, we are proud to share a novel kinase-focused dataset for FastGrow that features over 51,000 extension entries. Based on the previously reported Hinge Binder Collection, this set contains computationally validated fragments derived from reported bioactive compounds. Furthermore, ring systems were decorated with amine and amide groups to reflect features observed in approved drugs and investigated tool compounds. To capture full coverage of the shape volume, attachment points were placed on each ring system position and on connecting amine groups.
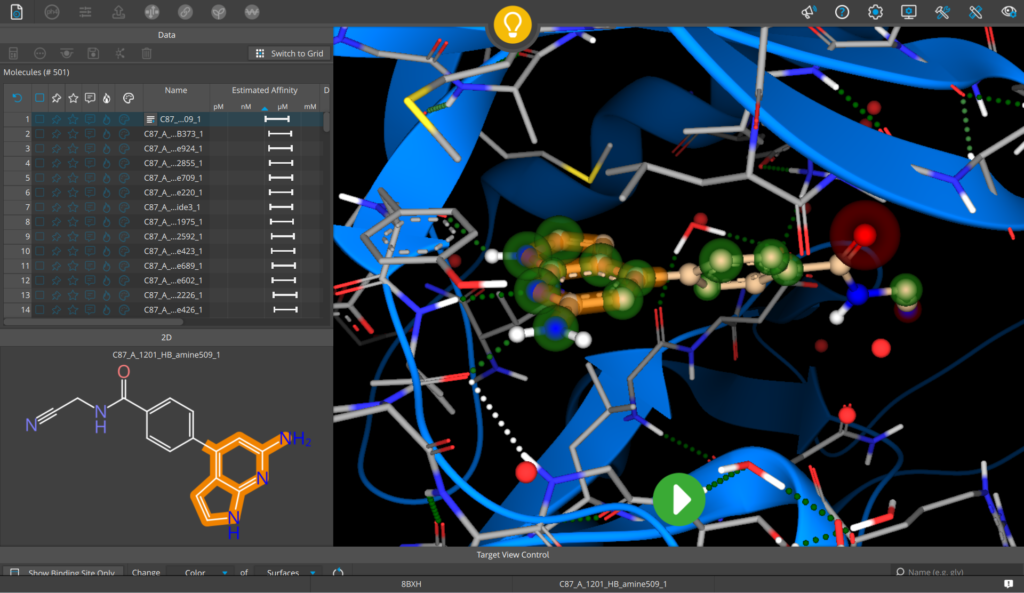
Application example of the new library on PDB 8BXH (JAK2 in complex with momelotinib). 500 results replacing the phenylmorpholinyl-bearing pyrimidine of momelotinib were generated in only a few minutes including the prediction of estimated binding affinity. Among 23 results scoring better than the co-complexed ligand, several additional potential interactions in the hinge binding region were observed, further expanding the medicinal chemistry tool box of the compound series.
The Hinge Binder library can be leveraged to mine for novel molecular motifs and therefore new compound leads. Identification of unique binding interactions could serve as the foundation for new drug series with improved physicochemical properties such as half time or penetration of the blood-brain-barrier. This is especially valuable when a project reaches a dead end, providing new paths for exploration even at a progressed discovery stage. Furthermore, finding novel chemical entities is crucial for securing patents, ensuring new compounds are protected and can be developed commercially without infringing on existing intellectual property.
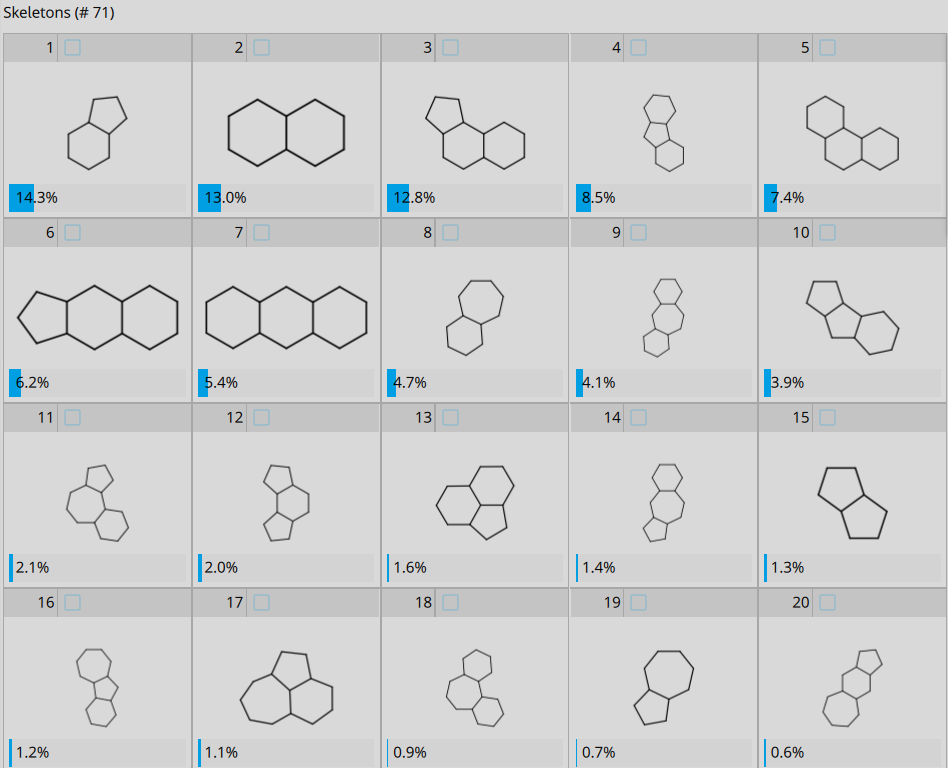
The Hinge Binder library features 71 different Bemis-Murcko molecular skeletons.
The Hinge Binder FastGrow library can also be used outside of kinase-focused projects to generate medchem-like compounds. Combining the library with pharmacophore constraints can efficiently screen for motifs satisfying particular interaction arrangements in a binding site.
Although the primary use of the library is structure-based prediction to identify the best candidates for follow-up, it can also be broadly applied to designing target-specific compound libraries. Incorporating methods such as molecular dynamics (MD) simulations or machine learning can further enhance the accuracy and efficacy of the predictions, providing deeper insights and more robust solutions.
The Hinge Binder library is offered free-of-charge and can be downloaded following this link.