More polished, more finesse, more efficiency – that’s version 14.1 of SeeSAR ‘Atlas’.
Once again, we’ve rolled up our sleeves to refine and tune the engines and gears of our drug design dashboard SeeSAR. While we were at it, we also put extra effort into new features that will take the user experience to the next level.
Expanding the Modeling Horizon with YASARA
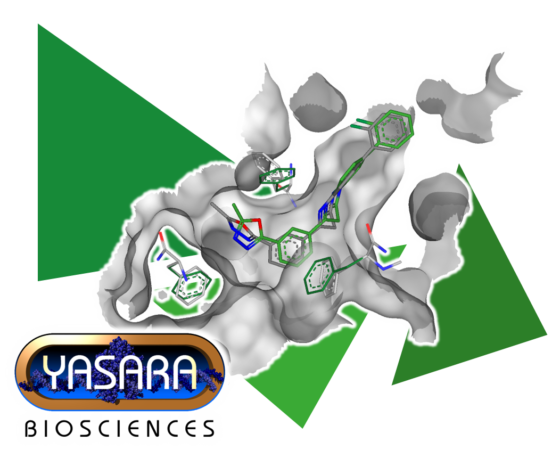
In our latest collaboration with YASARA, we are significantly expanding SeeSAR’s molecular modeling feature portfolio with a well-known and popular partner in drug discovery software solutions. The first feature to make its way into SeeSAR’s toolbox is the refinement of structures through energy minimization.
The application areas are particularly diverse: preparing a structure or model (for example from AlphaFold) for molecular modeling studies, docking setups, or entire virtual screening campaigns by eliminating possible torsional conformations and identifying the energetically most favorable arrangement of rotamers.
Additionally, it can be used to refine the poses of docked ligands, eliminating potential issues such as torsions or clashes, or to slightly adjust the size of the binding pocket to uncover new subpockets.
Our perspectives and potential application scenarios are summarized in this link.
Augmentations to Chemical Space Docking™
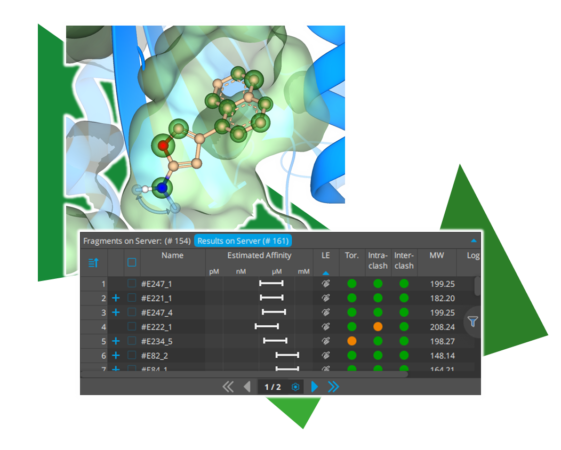
With the introduction of Chemical Space Docking™ in SeeSAR ‘Atlas,’ we have set the stage for the structure-based exploration of chemical space. To make the workflow even smoother and more efficient, several quality-of-life improvements have been added.
One innovation is the visual representation of possible extension vectors for relevant functionalities. As shown in the image above, for example, sp2 hybridized nitrogen atoms now indicate both directions towards which an extension can occur. This visual representation not only reflects the chemically correct steric behavior of the resulting compounds but also emphasizes the growth directions for easier assessment during the assessment of the poses.
Furthermore, ligand efficiency (LE) has now been set as the default parameter for sorting Chemical Space Docking™ results. Especially for the smallest fragments, it come handy to exploit when these exhibit an appropriate LE to enhance the quality of the results. Of course, it is still possible to sort the results by the HYDE score to achieve a balanced mix of both perspectives.
Visual Differentiation of Pharmacophore Constraints
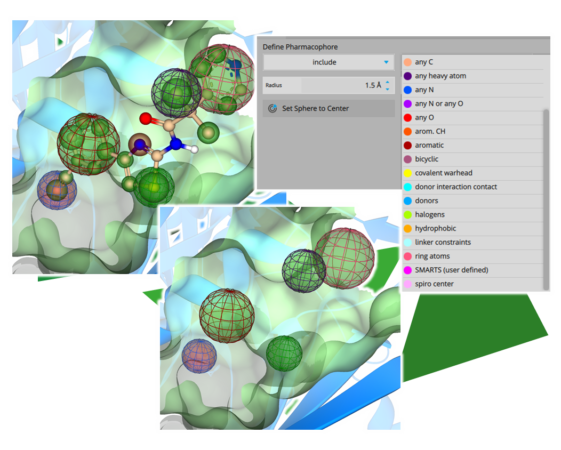
Another highlight is the representation of pharmacophore constraints. The placed spheres now visually reflect the chemical properties of the pharmacophore through color coding and indicate, with green and red coloring, whether they represent inclusive or exclusive filters.
Every mode in which constraints may be set (Docking, Space Docking, Inspirator, Analyzer, Similarity Scanner) uniformly supports this feature.
SeeSAR ‘Atlas’ 14.1
These were the highlights of SeeSAR 14.1. We are very excited for all the features, including the ones mentioned, as well as the small little ones that can be found in the full changelog. Feel free to download and test SeeSAR to experience the augmentations first hand!
Read the full changelog following this link.





