R group search is now supported in the Motif Matcher of infiniSee 6.1. Among several upgrades and augmentations, users will find this functionality to be the most convenient when it comes down to precisely navigating in ultra-large Chemical Spaces. By adding one or more R groups to a molecule in the Motif Matcher, users can now perform a screening to extract only compounds that contain one or more heavy atoms in the designated positions. The rest of the molecule will be left untouched.
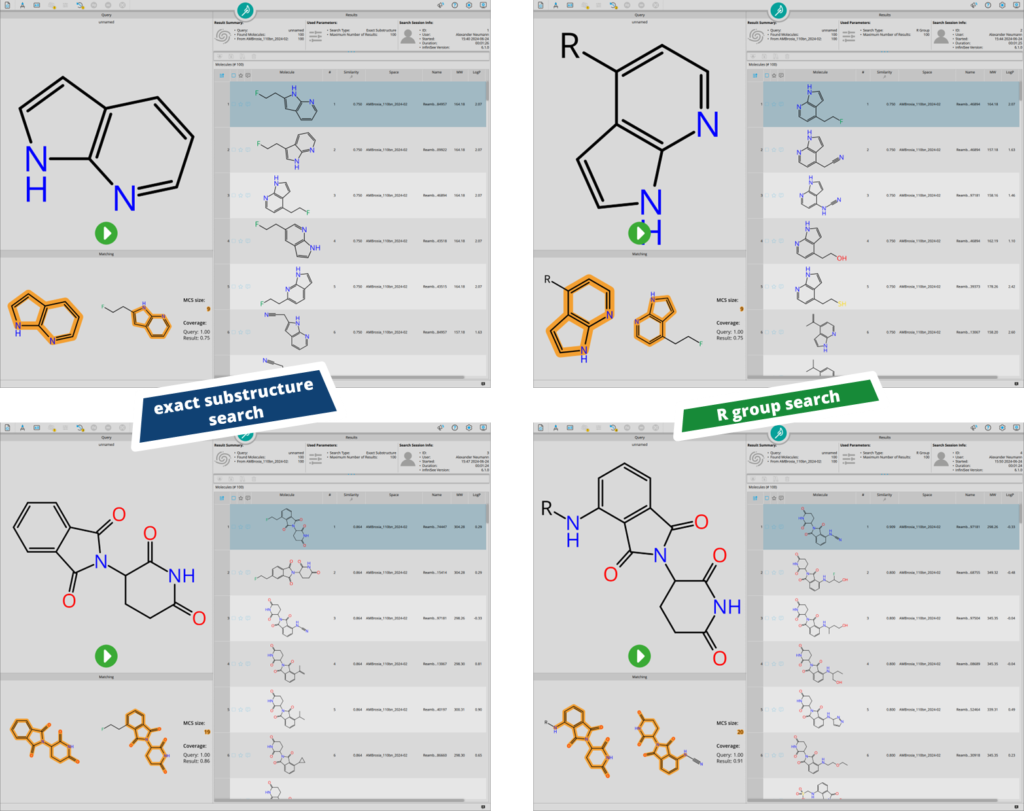
Differences between ‘exact substructure search’ and ‘R group search’. During exact substructure searching, all positions bearing an hydrogen atom are explored for decorations. During R group search, only desired growth directions of the scaffold are explored.
In the example above two different scenarios were explored. The first query features an azaindole (e.g. as a starting point for kinase inhibitor design) and the other features thalidomide (e.g. to mine for potential building blocks for molecular glues and PROTACS). Both cases highlight how the R group search can be extremely valuable during fragment-based drug discovery campaigns, where the direction of the extension vector is important to avoid clashes with the target’s binding site.
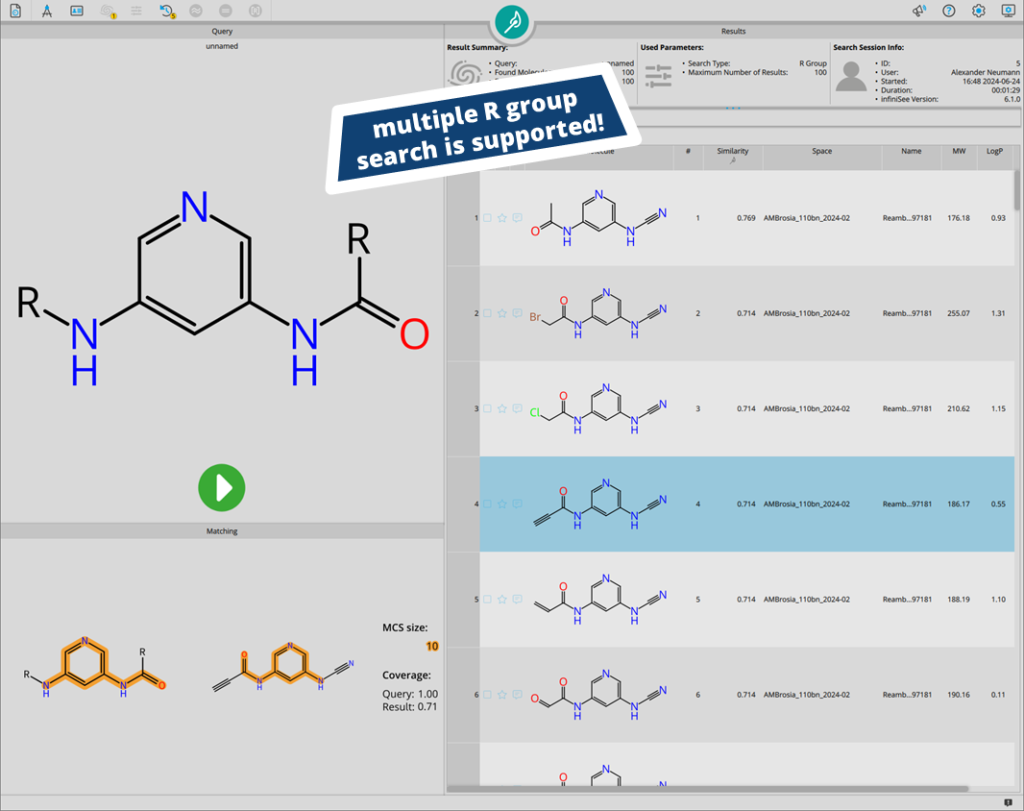
Users are not limited to the placement of a single R group during the search. If several extensions need to be explored, multiple growing extensions can be set up.
The second big addition is the introduction of ad hoc libraries to infiniSee. After performing a search in infiniSee, results can be bundled into an ad hoc library, that is saved in the background of infiniSee. This library can then be used as a screening source to perform subsequent searches for cross-validation of results, and orthogonal screening.
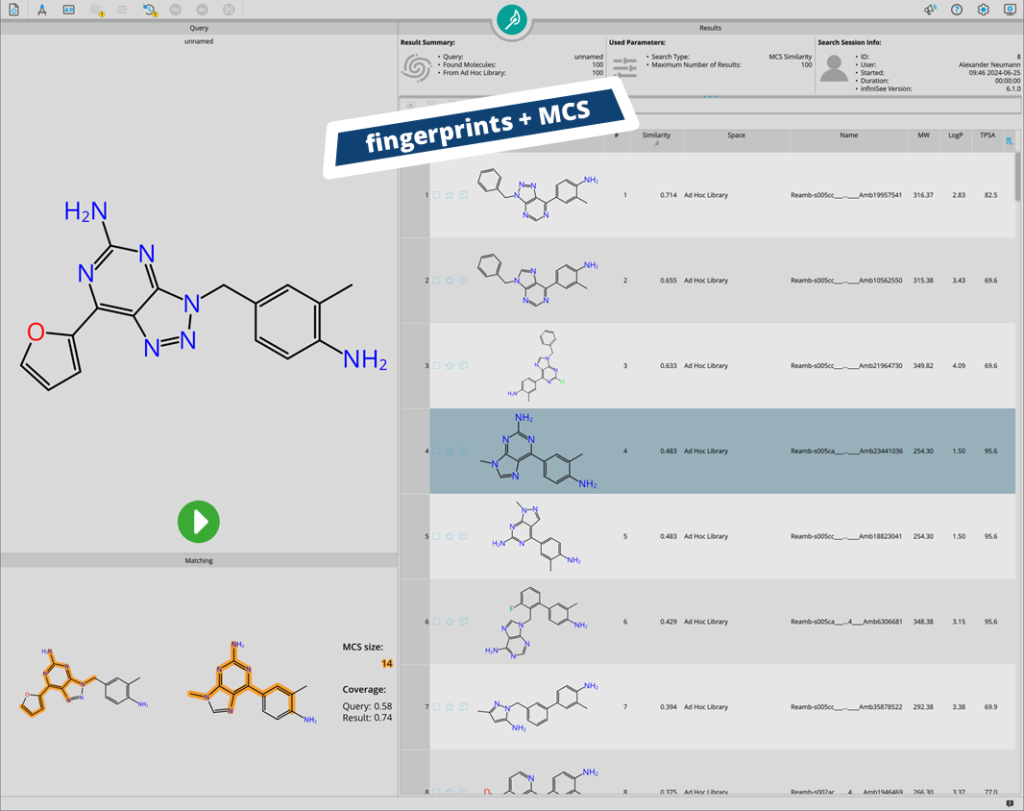
Ad hoc libraries are a convenient solution for similarity enrichment. In this case, results from the Analog Hunter (based on molecular fingerprints) are re-scored in the Motif Matcher (based on maximum common substructure matching).
We are excited to see how these additions to infiniSee will augment your drug discovery efforts and improve your experience navigating ultra-large Chemical Spaces.
We focused on two new major features of infiniSee 6.1. You can read the full changelog of infiniSee 6.1 following this link.
The update also entails:
- Improvement of distinct similarity columns in the Analyzer Mode.
- Expansion of supported chemical elements (Ge, As, Sb, Te)
- Additional visualization possibilities
- Easier reuse of queries
- Improved license status identification





