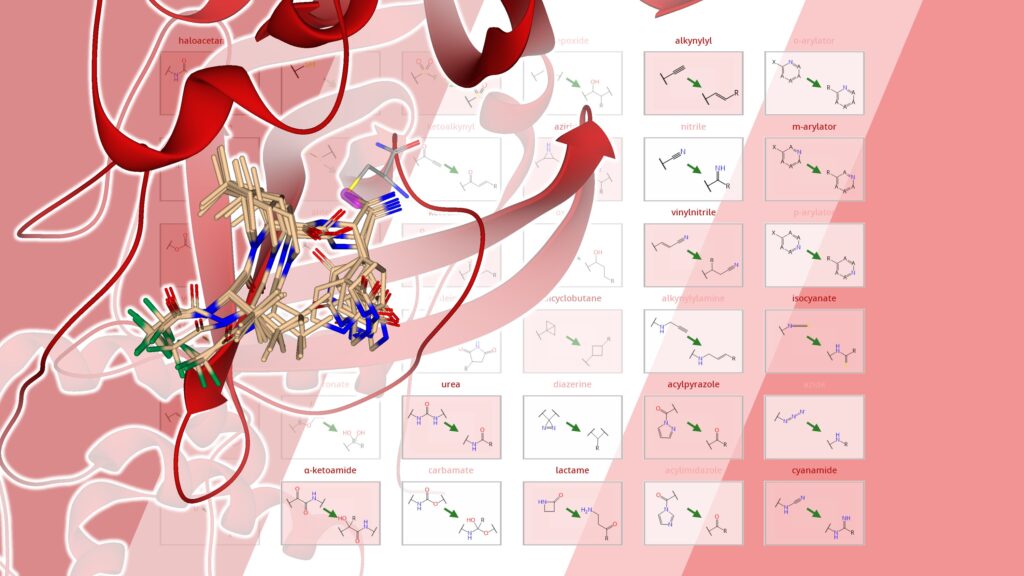
Covalent warhead transformation is a highly requested feature that found its way into the SeeSAR ‘Midas’ 13.1 update. Covalent warheads[1],[2] are privileged functional groups that can form reversible or irreversible bonds with nucleophilic residues in a binding site upon binding. As part of the covalent docking upgrade, potential covalent warheads are detected during the process and transformed into their target-bound form. For this, we have collected 36 most prominent molecular motifs and embedded the translation into the software.
During the docking process in SeeSAR, a covalent warhead is transformed into its target-bound form. The pose generation and subsequent scoring of the complex is a method than can be applied for SAR investigation and virtual screening of commercial libraries.
As of today, 36 covalent warheads are automatically detected and transformed during a covalent docking run:
-
- haloacetamide
- acrylamide
- acrylester
- vinylsulfones
- nitroalkenes
- nitroalkanes
- thioles
- disulfides
- aldehyde
- boron
- boronate
- α-ketoamide
- sulfonyl fluoride
- ketoalkynyl
- ketoamine
- maleimide
- urea
- carbamate
- epoxide
- aziridine
- oxetane
- bicyclobutane
- diazerine
- lactame
- alkynylyl
- nitrile
- vinylnitrile
- alkynylylamine
- acrylpyzarole
- acrylimidazole
- (o-, m-, p-)arylators
- iscocyanate
- azide
- cyanamide
The image below displays how the covalent warhead is transformed during the process.
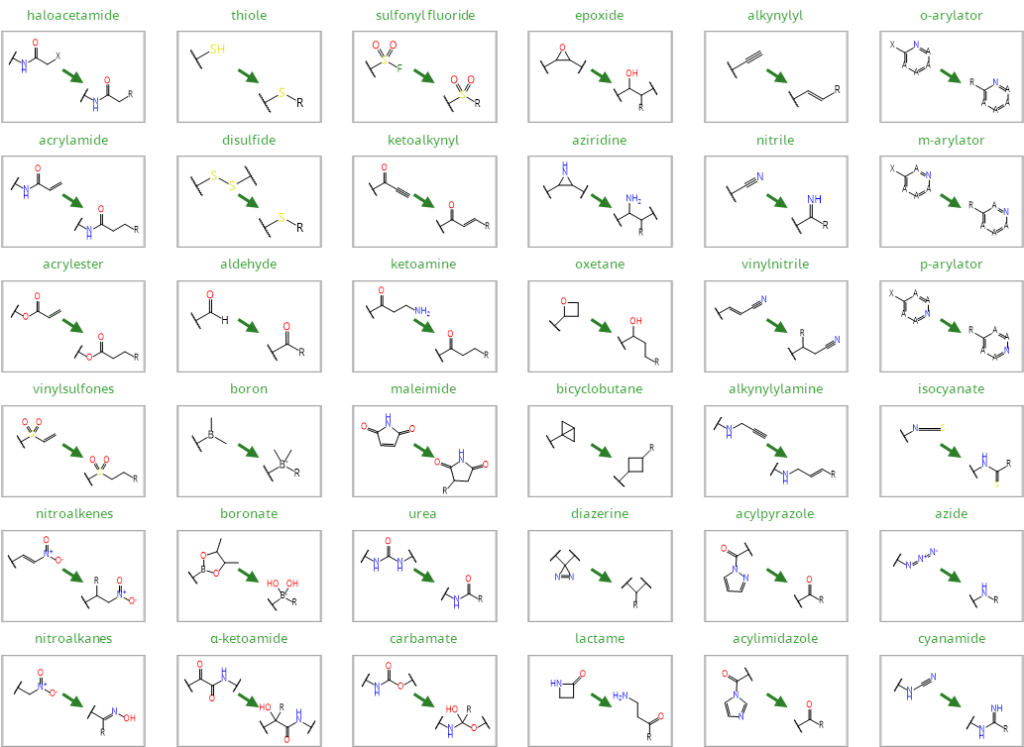
Most frequently used covalent warheads in drug discovery that are automatically detected in SeeSAR. The figure shows the transformation of the warheads into their target-bound form.
With the covalent docking update, following residues can conveniently be selected in the Docking Mode as target: Cys, Ser, Thr, Lys, Tyr, Asn, Gln. Yet, it is still possible to introduce an attachment point (so-called ‘linker’) to any part of the target structure (even if it is an DNA or RNA, or a co-complexed ligand/cofactor) in the Protein Editor Mode to perform covalent docking at any part of the complex. After adding the linker, re-define the binding site and select your introduced attachment point in the Docking Mode for the generation of poses.
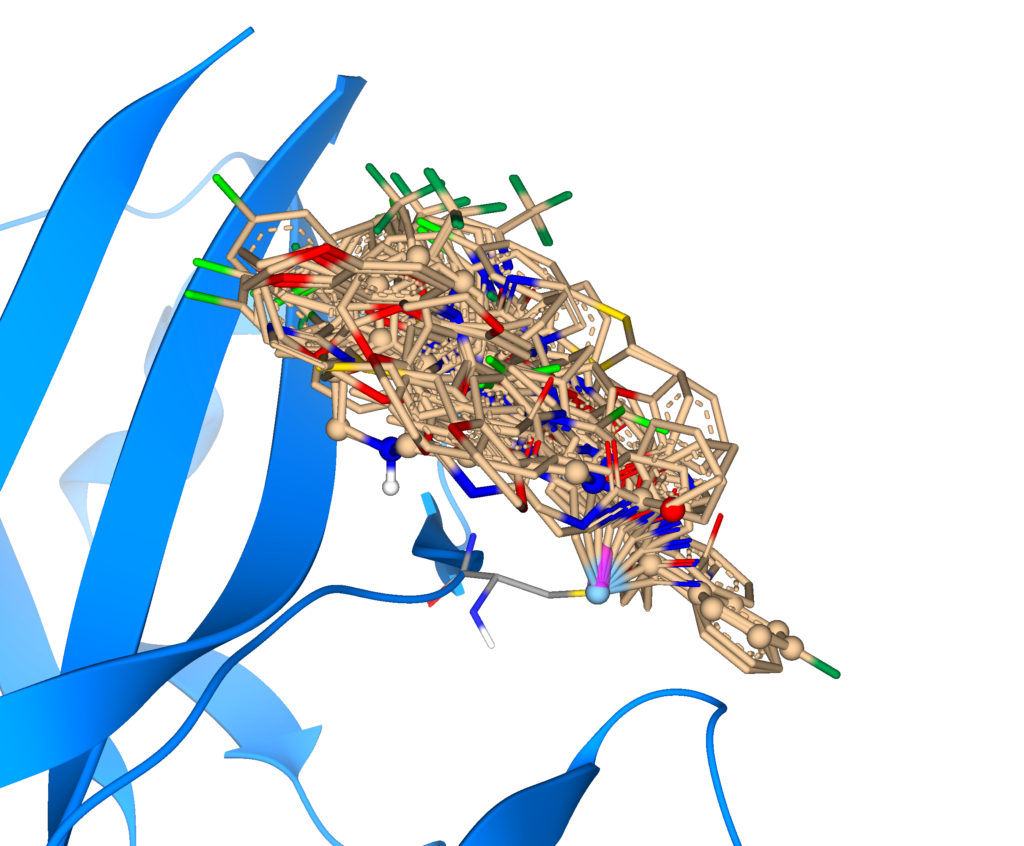
If a stereo center is introduced during docking, all resulting enantiomer pairs are sampled during the process.
During the docking process, all resulting enantiomers of the ligand-introduced bond are sampled to cover all possibilities of the binding event outcome. Furthermore, users can optimize their dockings with additional settings: Per default, the terminal bond of the residue side chain’s heavy atom is sampled during the docking. If a particular angle is requested for the run, users can decide to keep it rigid in the docking settings.
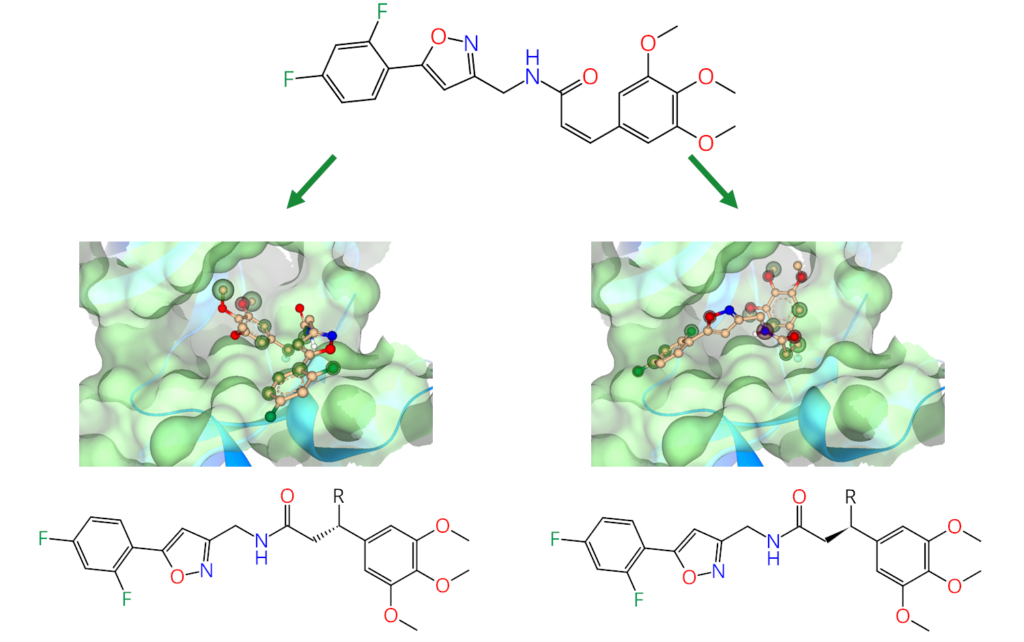
Furthermore, users can apply ‘covalent warhead’ pharmacophore constraints during standard docking. This definition features the previously mentioned residues and identifies the covalent warhead using a SMARTS definition. By doing this, it becomes possible to sample a set of ligands for their potential to bind near the target residue before forming the covalent bond, providing an additional layer of cross-validation for their interactions with the target structure. This approach can enhance the identification of potential active compounds during a virtual screening campaign.
Take your ligand containing a covalent warhead and test out the feature in SeeSAR yourself. Simply download the software following this link and get started!