The hinge binder motif is a prominent molecular feature present in the majority of kinase inhibitors, a class of compounds acting at one of the most important drug targets. So far, over 70 small molecules have received approval by the FDA for the treatment of many diseases, including cancer, by regulating the activity of two dozen distinct protein kinases. Here, we unveil a chemically diverse set of computationally assessed hinge binder fragments for your in-house workflows and projects.
Kinases — How to Drug Them
The most prominent mechanism of action of kinase inhibitors is their direct competition with ATP in the hinge binding domain that connects two domains of the protein. Once blocked by the inhibitor, the kinase loses its ability to transfer phosphate groups from ATP to other molecules, resulting in disruption of the kinase activity ultimately leading to the modulation of various cellular processes.
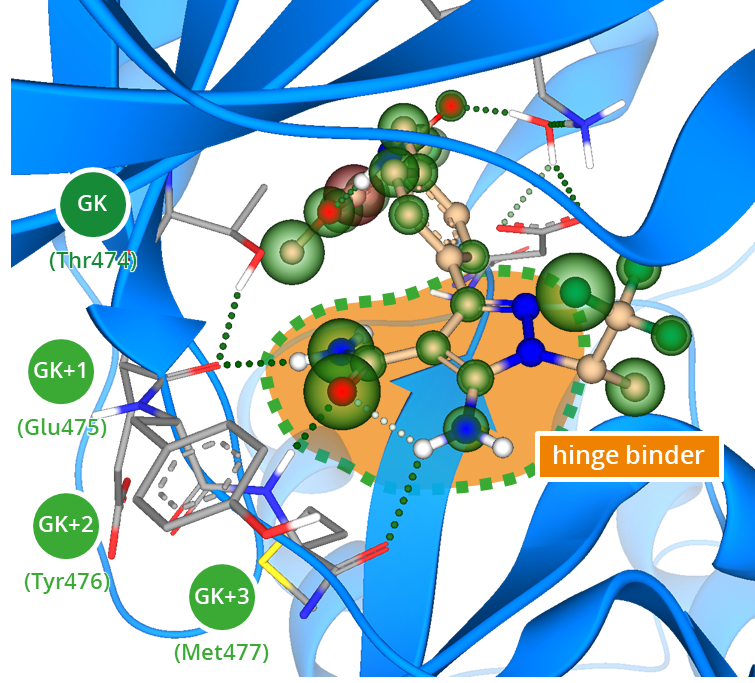
Complex of Bruton’s tyrosine kinase (BTK) bound to pirtobrutinib (PDB ID: 8FLL). The hinge binding motif is composed of three amino acids following the gatekeeper residue (GK). Efficient hydrogen bond interactions with the backbone stabilize the kinase inhibitor in the binding cleft.
The hinge binder in a kinase inhibitor mimics the interaction pattern of ATP’s nucleobase, establishing hydrogen bonds with the backbone of residues adjacent to the gatekeeper (GK). Typically, a ligand’s hydrogen bond acceptor engages with the NH of the third residue following the gatekeeper (GK+3). Additional hydrogen bond interactions may also occur with the carbonyl of GK+1 or GK+3, allowing for up to three potential engagements with the protein backbone. Several molecular scaffolds have been identified for their effective alignment with the hinge binding region through their 3D pharmacophore arrangement. Ultimately, the development of novel drug candidates requires innovative intellectual property (IP) to address common selectivity issues among related kinases and their subtypes.
Hinge Binder Diversity Set
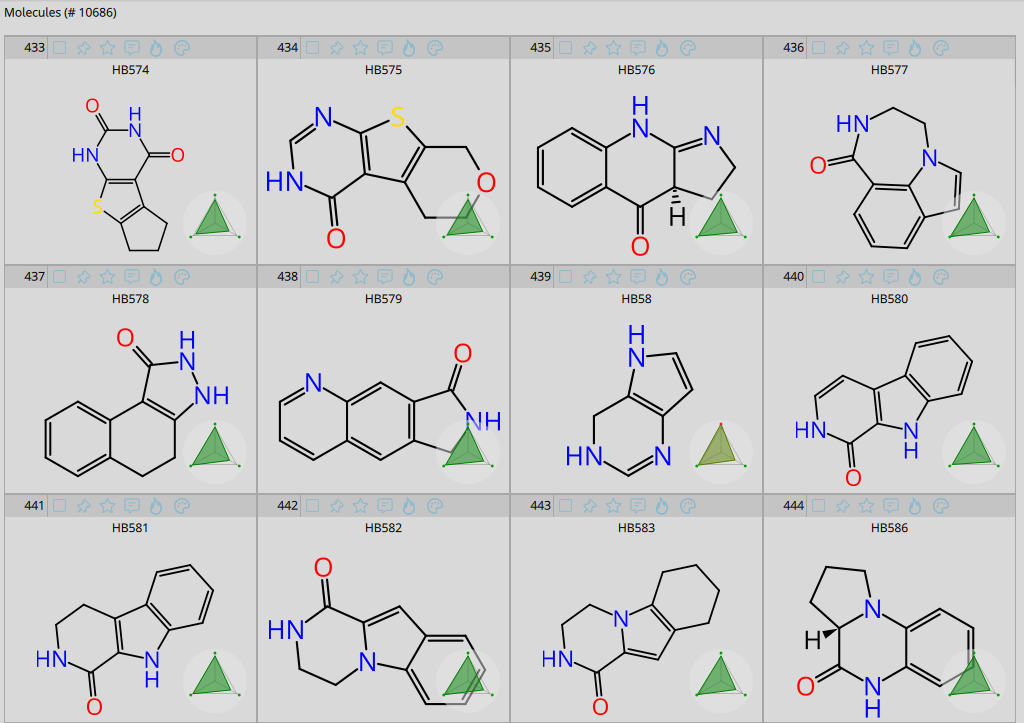
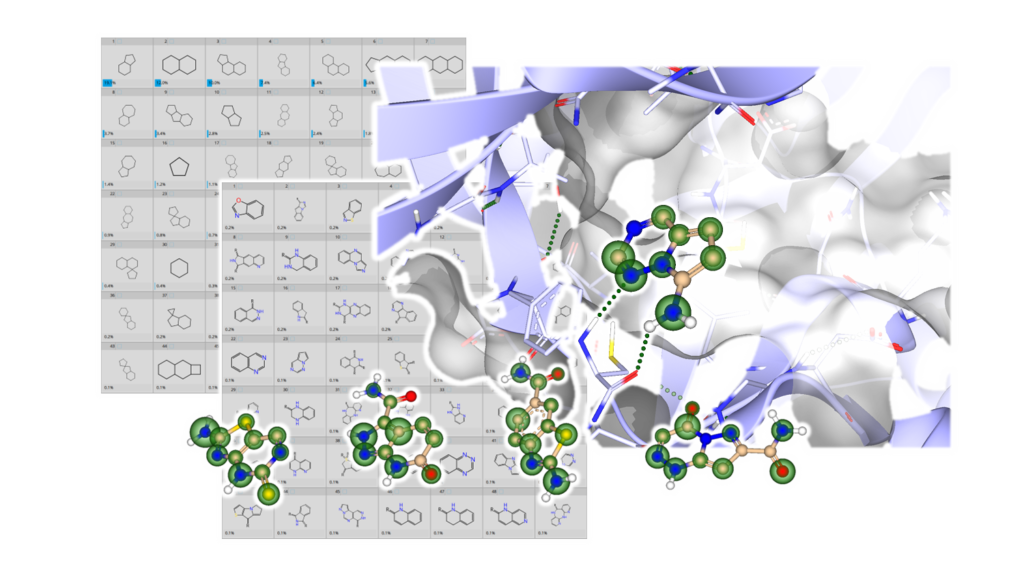
Examples from the BioSolveIT Hinge Binder Diversity Set.
As a little treat for computational approaches and as a novel source for compound ideation, we have created a curated and diverse set of potential hinge binding fragments. For this, we have extracted bioactive molecules from the ChEBML database and isolated their molecular fragments. Since the presence of amino and amide groups on a molecular fragment have been observed to be important for binding at the hinge region in some cases, we have diversified and augmented the collection by adding both such groups to unoccupied positions of the ring system. Subsequently, the fragments were assessed for their hinge region binding ability via docking at distinct kinases. This was done with our visual drug discovery dashboard SeeSAR. We also applied pharmacophore constraints to ensure proper interactions with NH of GK+3 and potential interactions with the carbonyl groups of GK+1 and GK+3. Only fragments that would potentially bind to at least 3 kinases, and contained a maximum of three rings, were included in the final Hinge Binder Diversity Set: resulting in 10,686 computationally validated compounds.
Versatile Application Possibilities
This set can be used to jump-start your kinase targeting journey! Here are a few ideas:
- Dock the fragments into the hinge binding region to select the most promising candidates for follow up.
- Use the fragments for scaffold hopping approaches, or to improve ADME properties.
- Combine them with the pocket exploration tool FastGrow to efficiently expand your fragments and complement the binding site.
- Use the database as an excellent source to screen for fragments that may display selectivity towards your kinase subtype.
- Exploit the fragments for machine learning and artificial intelligence approaches.
You can download the Hinge Binder Diversity Set following this link (10,686 compounds, SMILES).
Should you have any questions about the Hinge Binder Diversity Set, please reach out to us via support@biosolveit.de.